Functions and Iterations
Learning to do repeated tasks in R
Meenakshi Kushwaha
18th August, 2022
Functions
When should you write function?
You should consider writing a function whenever you’ve copied and pasted a block of code more than twice (i.e. you now have three copies of the same code).”
- R for Data Science
Example of a repeated task
In the gapminder dataset calculate the range of population, gdp, and life expectancy
Your first function
Test-run your function
What did we need?
- name of the function
max_minus_min
- what do you want the function to do? OR body
round((max(x) - min(x)), 1)
- what does the function need to work? OR ingredient
x
A function returns the last statement it evaluates
Quiz
What will the following code return?
- 10
- 20
- 30
- Will give an error
How to re-use your function
- Step 1: Save your function as
.Rscript - Step 2: Source the script using
Demo
Good Practices
- descriptive names, use verbs for function names
- use snake case in names
- be consistent with naming
- use common prefix for a family of related functions
Other tips
- Use comments for #why. Code should explan the #how
- Shorcut (extract function)
Iterations
map() Function
FOR EACH ____ DO _____
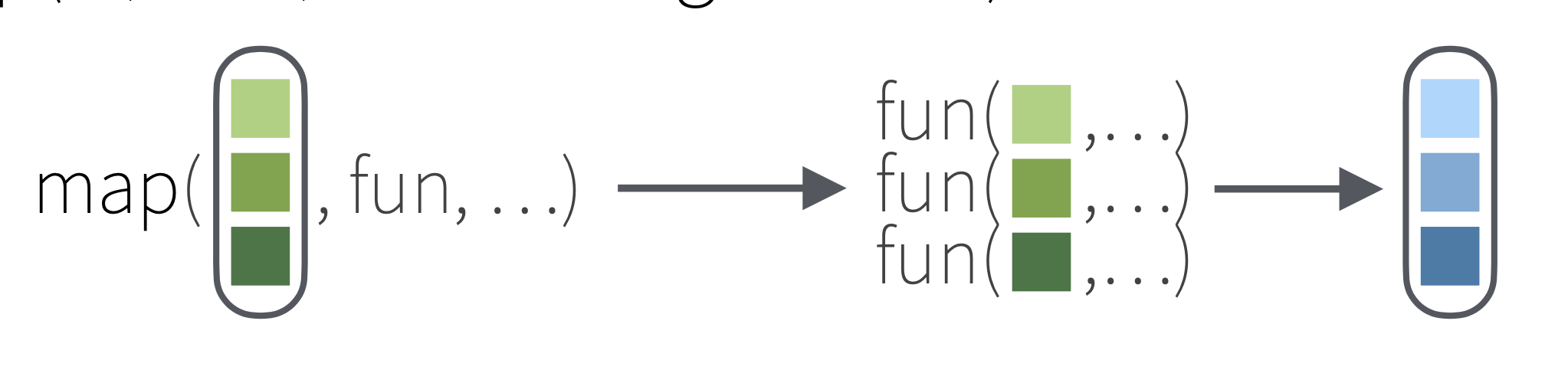
map(.x, .f)
.x is a vector/list/dataframe
.f is the action that you want to do with each element of .x
Types of map() functions
map(.x, .f) is the main mapping function and returns a list
map_df(.x, .f) returns a data frame
map_dbl(.x, .f) returns a numeric (double) vector
map_chr(.x, .f) returns a character vector
map_lgl(.x, .f) returns a logical vector
Input to map() functions
- a vector (of any type)
- in which case the iteration is done over the entries of the vector
- a list
- in which case the iteration is performed over the elements of the list
- a data frame
- in which case the iteration is performed over the columns of the data frame
Example
Apply class() function to each column of gampminder data
$country
[1] "factor"
$continent
[1] "factor"
$year
[1] "integer"
$lifeExp
[1] "numeric"
$pop
[1] "integer"
$gdpPercap
[1] "numeric"Example
What is the number of distinct values in each column
country continent year lifeExp pop gdpPercap
142 5 12 1626 1704 1704 Example
What is the median of all numeric columns?
year lifeExp pop gdpPercap
1979.5000 60.7125 7023595.5000 3531.8470 When things get complicated, use ~
Sometimes you may want to define your own function and apply to each column
function(x) { x + 10 }
can be replaced with ~{. + 10}
Using ~ with map functions
Quiz
What is the output of the following code?
Using ~ with map functions
country continent year lifeExp pop gdpPercap
142 5 12 1626 1704 1704 Using ~ with map functions
Fitting a linear model with different groups of the data
$Africa
Call:
lm(formula = lifeExp ~ pop, data = df)
Coefficients:
(Intercept) pop
4.816e+01 7.150e-08
$Americas
Call:
lm(formula = lifeExp ~ pop, data = df)
Coefficients:
(Intercept) pop
6.353e+01 4.587e-08
$Asia
Call:
lm(formula = lifeExp ~ pop, data = df)
Coefficients:
(Intercept) pop
5.992e+01 1.901e-09
$Europe
Call:
lm(formula = lifeExp ~ pop, data = df)
Coefficients:
(Intercept) pop
7.162e+01 1.650e-08
$Oceania
Call:
lm(formula = lifeExp ~ pop, data = df)
Coefficients:
(Intercept) pop
7.207e+01 2.545e-07 Using ~ with map functions
Fitting a linear model with different groups of the data
$Africa
Call:
lm(formula = lifeExp ~ pop, data = .)
Coefficients:
(Intercept) pop
4.816e+01 7.150e-08
$Americas
Call:
lm(formula = lifeExp ~ pop, data = .)
Coefficients:
(Intercept) pop
6.353e+01 4.587e-08
$Asia
Call:
lm(formula = lifeExp ~ pop, data = .)
Coefficients:
(Intercept) pop
5.992e+01 1.901e-09
$Europe
Call:
lm(formula = lifeExp ~ pop, data = .)
Coefficients:
(Intercept) pop
7.162e+01 1.650e-08
$Oceania
Call:
lm(formula = lifeExp ~ pop, data = .)
Coefficients:
(Intercept) pop
7.207e+01 2.545e-07 Strategy
- Solve for one element
- Turn it into a recipe
- Use
map()to solve for all elements
Demo
Reading multiple files with purrr package
Reading multiple files
We have data from different countries in seperate .csv files that we need to combine in a single dataset for analysis
Reading multiple files
Step 2
Make a list of all the files
[1] "/Users/meenakshikushwaha/Dropbox/R projects/github/cstep-iterations/data/china_gm.csv"
[2] "/Users/meenakshikushwaha/Dropbox/R projects/github/cstep-iterations/data/india_gm.csv"
[3] "/Users/meenakshikushwaha/Dropbox/R projects/github/cstep-iterations/data/japan_gm.csv"
[4] "/Users/meenakshikushwaha/Dropbox/R projects/github/cstep-iterations/data/nepal_gm.csv"Reading multiple files
Step 3
Read and combine all files using map_dfr()
# A tibble: 16 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 China Asia 1952 44 556263527 400.
2 China Asia 1957 50.5 637408000 576.
3 China Asia 1962 44.5 665770000 488.
4 China Asia 1967 58.4 754550000 613.
5 India Asia 1952 37.4 372000000 547.
6 India Asia 1957 40.2 409000000 590.
7 India Asia 1962 43.6 454000000 658.
8 India Asia 1967 47.2 506000000 701.
9 Japan Asia 1952 63.0 86459025 3217.
10 Japan Asia 1957 65.5 91563009 4318.
11 Japan Asia 1962 68.7 95831757 6577.
12 Japan Asia 1967 71.4 100825279 9848.
13 Nepal Asia 1952 36.2 9182536 546.
14 Nepal Asia 1957 37.7 9682338 598.
15 Nepal Asia 1962 39.4 10332057 652.
16 Nepal Asia 1967 41.5 11261690 676.Using map_dfr()
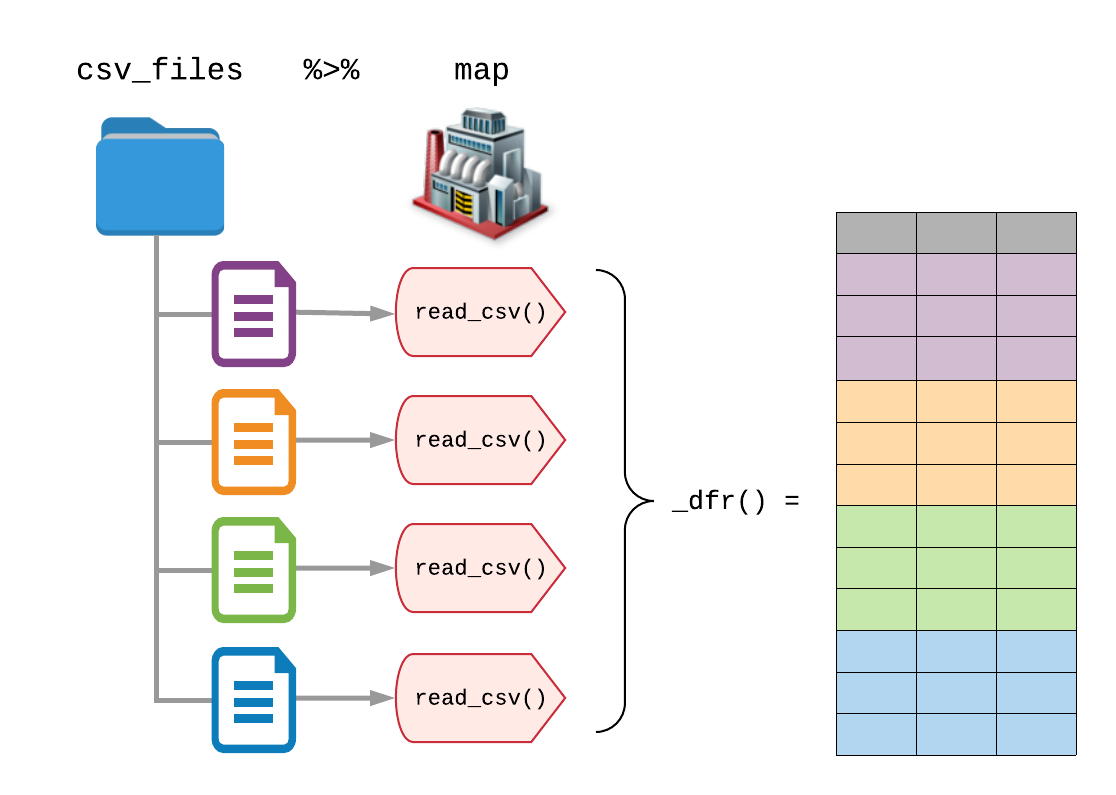
The additional _dfr() tells purrr to return a data frame (df) by row-binding each element together (r)
Source: Blog by Garrick Aden-Buie